Paper Review - UNet
[Reference] UNet Paper
Abstract.
The U-Net, proposed in 2015, represents a CNN framework primarity employed for image segmentation tasks. this model was designed for medical image segmentation.
It comprises two main features:
- Contracting path: Responsible for feature extraction of context(Overall image with information such as texture, color, shape, etc. around a pixel or object)
- Symmetric expanding path: Facilitating precise localization
Due to these features, U-Net demonstrates remarkable performance in segmentation tasks and outperforms the prior best method(a sliding-window convolutional network)
- Sliding-window convolutional network
In order to find multi objects in a large image, divide the entire image into areas of the right and apply the localization network made in the previous step repeating for each area.
U-Net is so fast because inference time of segmentation of a 512x512 image takes less than a second on a recent GPU. Just looking at it, the existing method sliding-window convolutional network seems to take a long time to infer.
1. Introduction
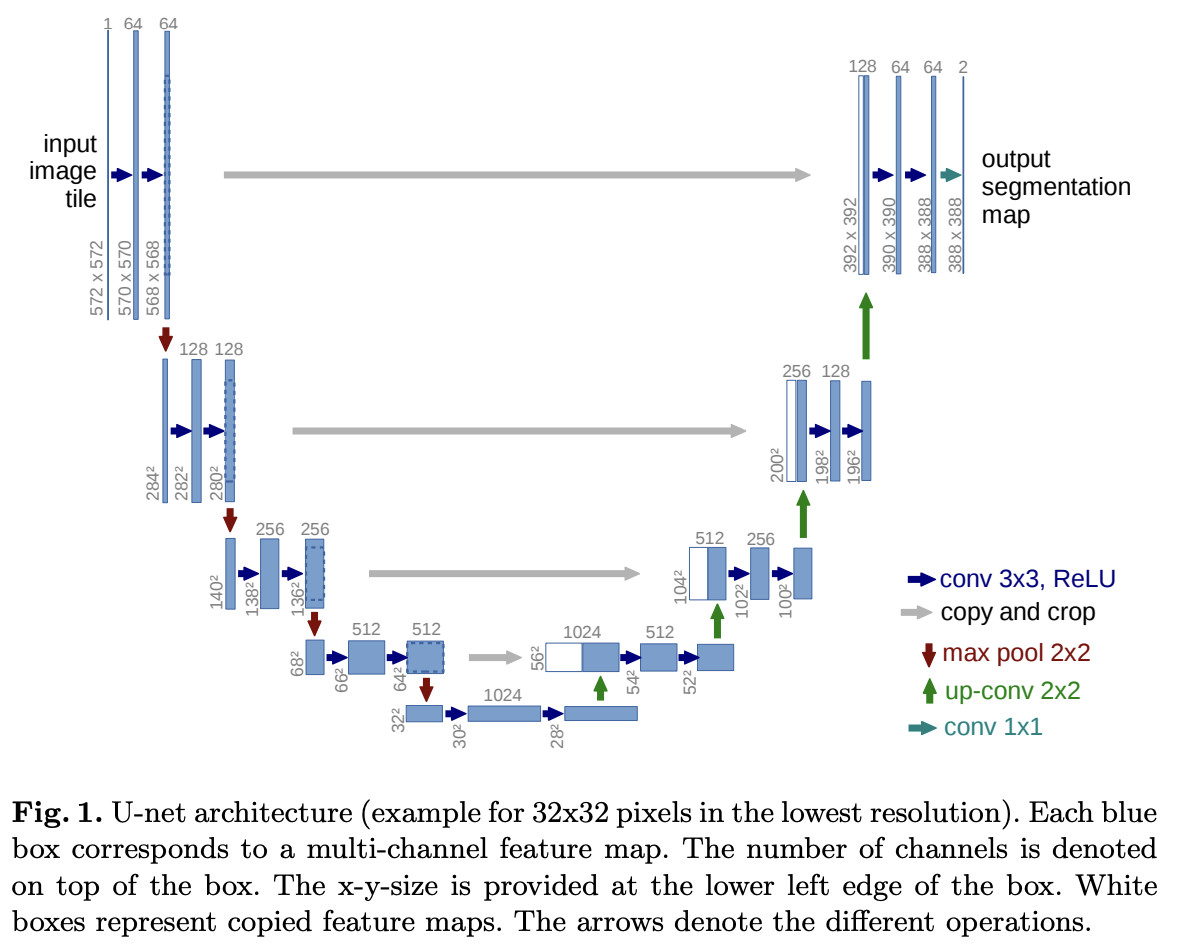
This model was built upon by FCN(you can check the organized FCN in that link). In this paper, they tried to modify and extent this model such that it works with very few training datasets and yields more precise result (see Figure 1).
One significant modification in this architecture is the inclusion of a large number of feature channels in the upsampling part. This enhancement enables the network to convey context information to higher-resolution layers. Consequently, the expansive path(=Decoding path) is approximately symmetric to the contracting path(=Encoding path), resulting in a U-shaped architecture.
Overlap-tile strategy
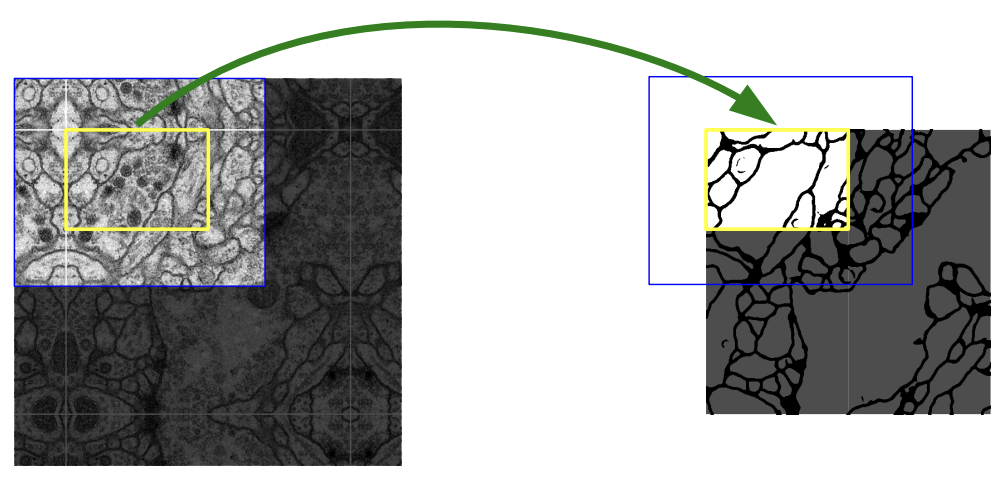
The blue box in above image (left) represents the input image to the network. Due to the use of valid convolutions (without any padding), the output is supposed to be the smaller yellow box (right).
They’re illustrating that the image they want to predict on is larger than the input to the network, possibly due to limited GPU memory. Hence, they need to perform inference multiple times using different parts of the input.
On the right side, imagine shifting the yellow box downward so that it aligns with the original square. Repeat this process to “tile” the output space. This requires a larger region of the input (blue) for inference. For non-overlapping yellow boxes in the output, overlapping blue boxes are needed for the input.
Mirroring extrapolation strategy
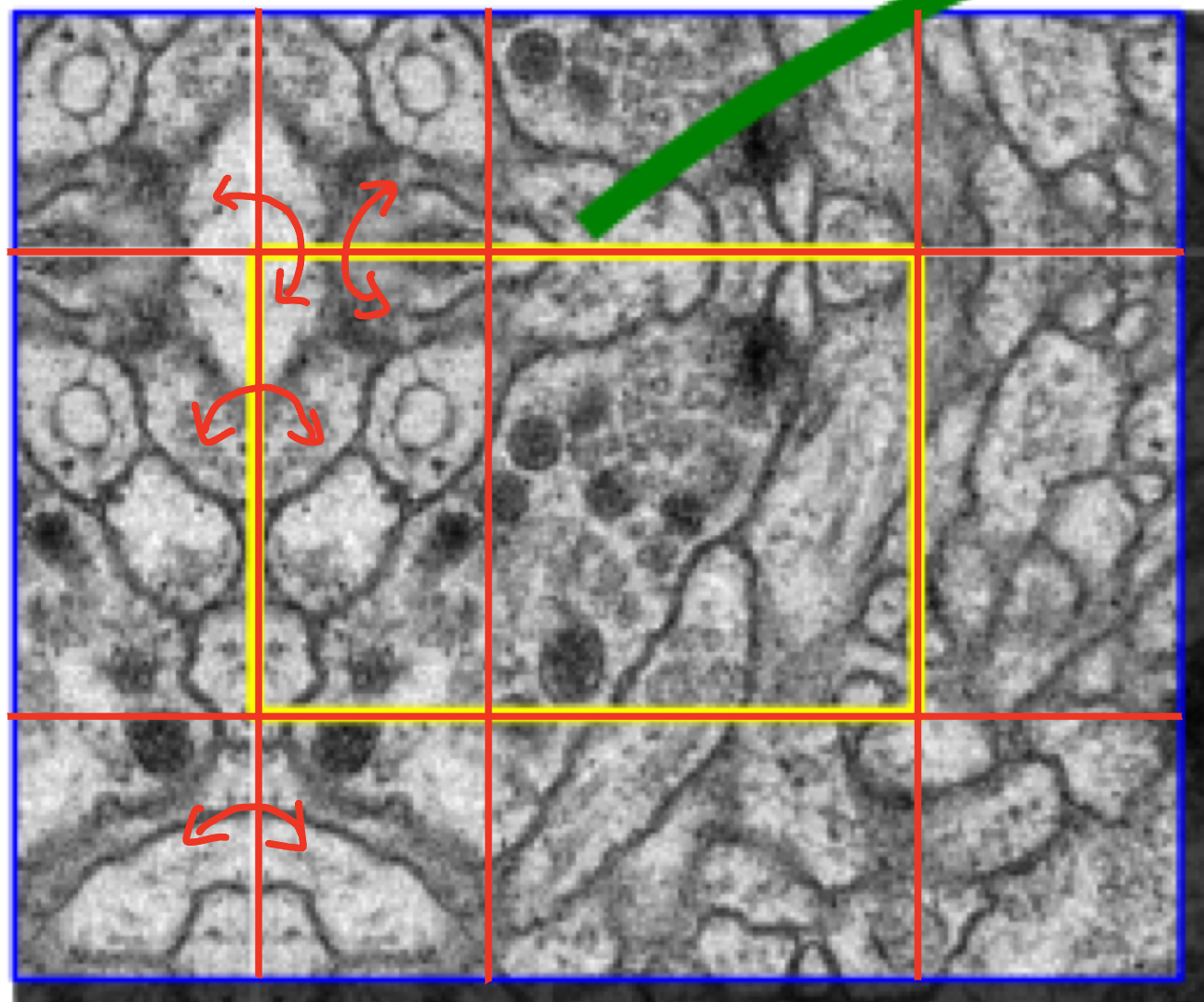
As you can see from the red arrow, it is a method of padding the empty space of the boundary by symmetrical existing images.
This strategy to improve predicion result about border regions of an imput image.
Elastic deformation
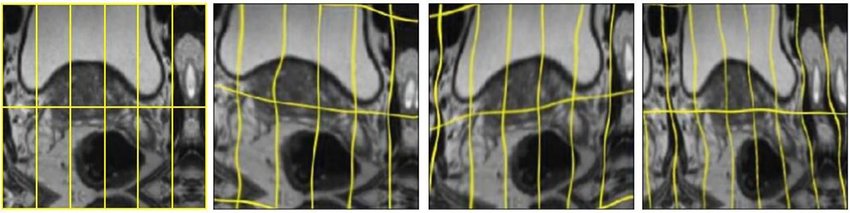
Elastic deformation is a technique used to augment training data by applying random distortions to images, helping AI models learn to recognize objects under different conditions (see above image).
This enables the network to learn invariant to such deformations, eliminating the necessity to encounter these transformations in the annotated image corpus. This practice is crucial in biomedical segmentation, where tissue deformation is a common variation, and realistic deformations can be efficiently simulated.
2. Network Architecture
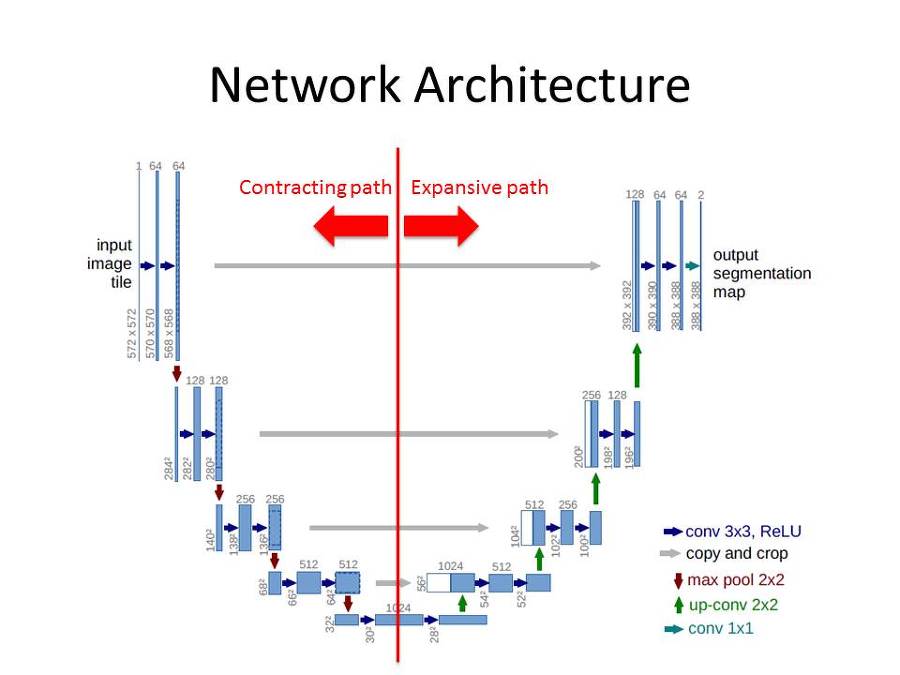
This network architecture can be divided into two main structures.
- Contracting path
- Two 3x3 convolution (unpadded convolution) + ReLU
- 2x2 max poolig operation with stride 2
- Double the number of channels when downsampled
- Expansive path
- 2x2 Transposed convolution that halves the num of feature channels
- Concatenation with the correspondingly cropped feature map from the contracting path
- Two 3x3 convolution (unpadded convolution) + ReLU
The last 1x1 convolution changes the number of 64 filters to the number of classes to be segmented
3. Training
- This model used SGD with momentum 0.99 as an optimizer
- To use of the GPU memory, they favor large input tiles over a large batch size
- Reduce the batch to a single image
- Pixel-wise soft-max
- $p_{k}(x)=exp(a_{k}(x))/(\sum_{k’=1}^{K}exp(a_{k’}(x)))$
- $k$ - Feature channel
- $a_{k}(x)$ - The activation in feature channel k at pixel position x
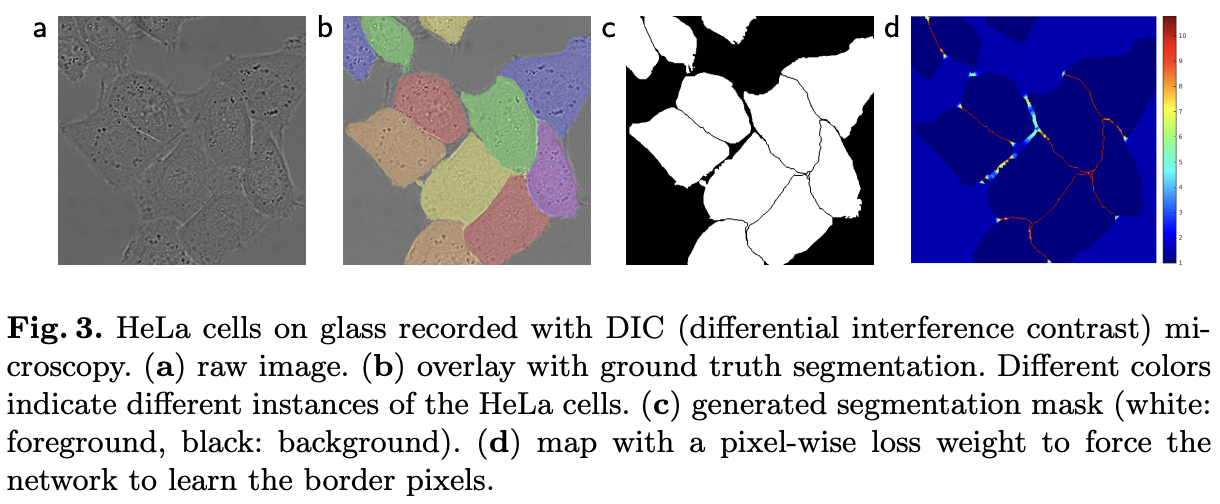
- Weight map was designed to give more importance to border pixels between touching cells (See Figure 3c and d)
- $w(x) = x_{c}(x) + w_{0}\cdot exp(-\frac{(d_{1}(x)+d_{2}(x))^{2}}{2\sigma ^{2}})$
- $w_{c}(x)$ - The weight map to balance the clas frequencies
- $d_{1}/ d_{2}$ - The distance to the border of the first, second nearest cell
- $w_{0} = 10$, $\sigma\approx5$ pixels
- Cross-Entropy loss function
- The loss value of each pixel was calculated by multiplying the pixel’s unique weight map with the cross-entropy.
- $E = \sum_{x\in\Omega}w(x)\log(p_{l(x)}(x))$
- Gaussian distribution initialization
- standard deviation = $\sqrt{\frac{2}{N}}$
- N = filter size x previous channel size
3.1 Data Augmentation
- Shift and rotation invariance
- Gray value
- Elastic deformation (I have already explained this technique above)
4. Experiments
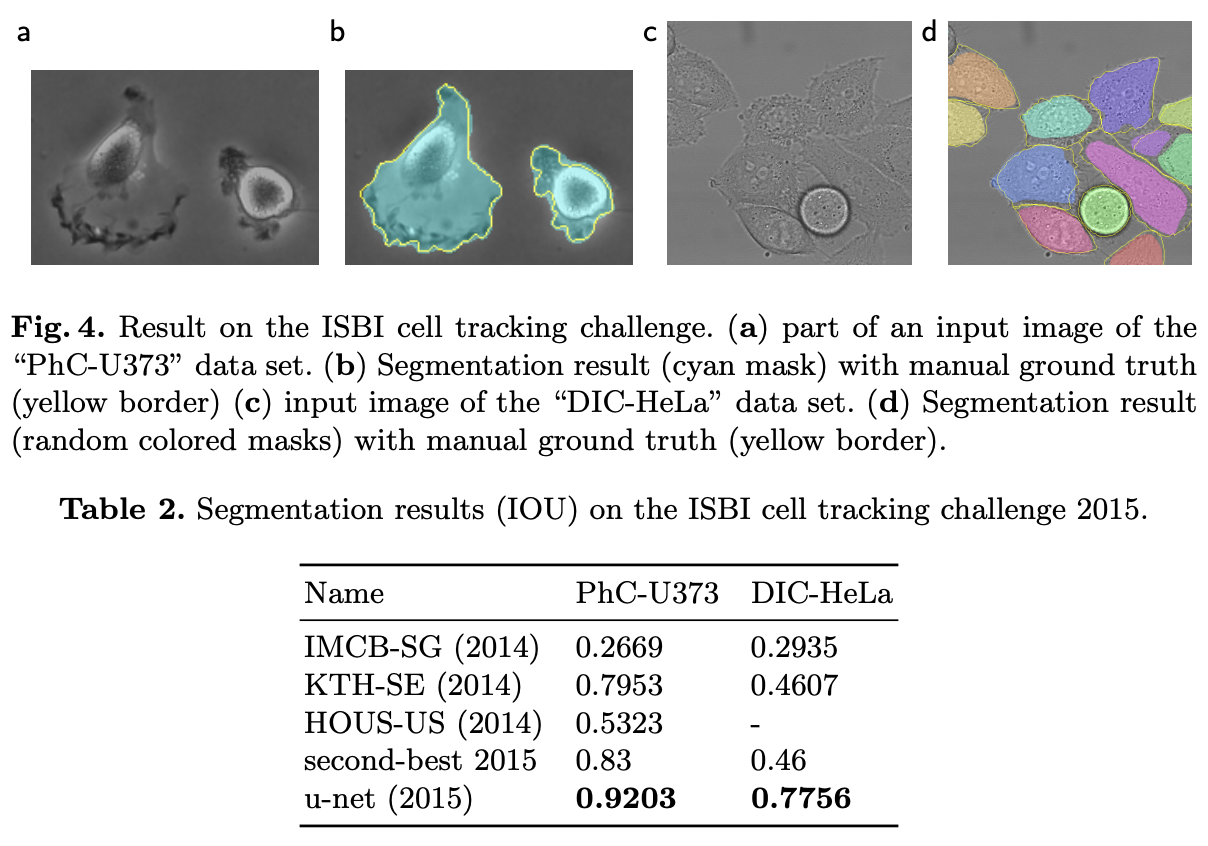
As evident from the results, UNet achieved SOTA performance with IOU scores of 92% and 77.5% on the two datasets, respectively.
Conclusion
In summary, the U-Net architecture offers exceptional performance in biomedical image segmentation tasks, leveraging techniques like data augmentation with elastic deformations to achieve accurate results with minimal annotated data. Its efficient training time, as demonstrated on an NVidia Titan GPU (8GB), makes it a versatile and promising solution for various biomedical imaging challenges.